PyDGC, a flexible and extensible Python library for deep graph clustering (DGC), is compatible with frameworks such as PyG and OGB. It supports the easy integration of new models and datasets, facilitating the rapid development, reproduction, and fair comparison of DGC methods.
- 🔥2025.07: API documentation is released.
- 🔥2025.07: PyDGC is now available on PyPI.
- 2025.05: Release source code of PyDGC.
Deep graph clustering, which aims to reveal the underlying graph structure and divide the nodes into different groups, has attracted intensive attention in recent years.
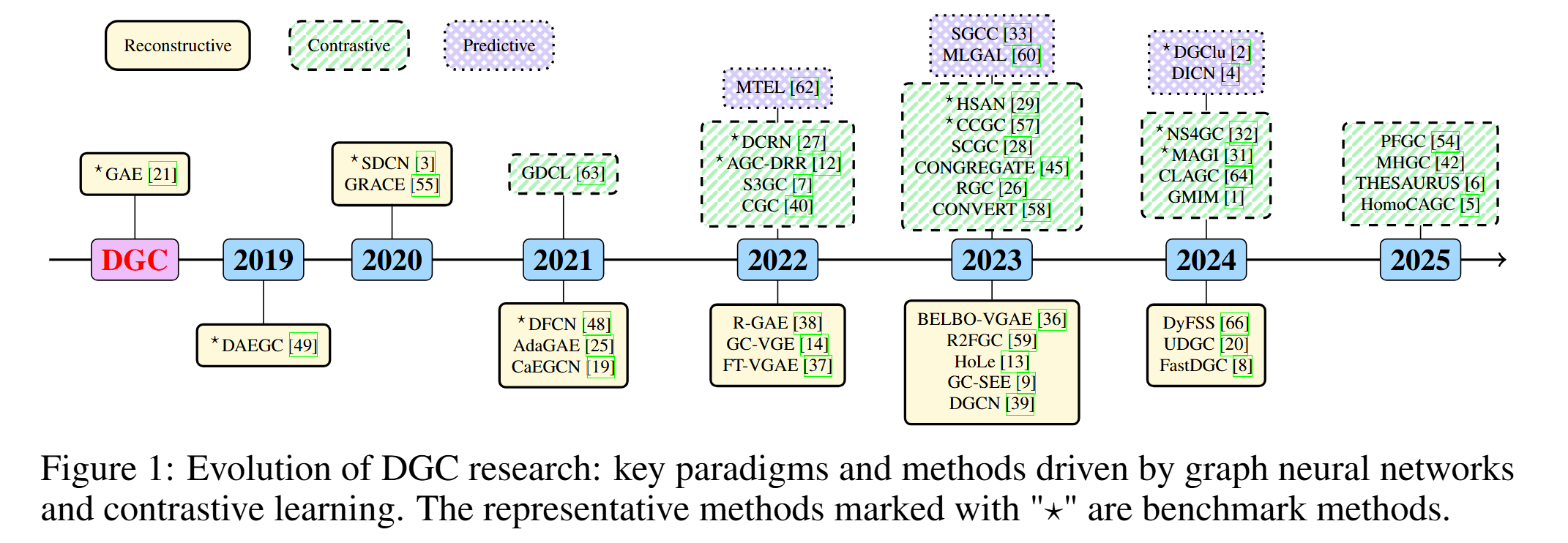
More details can be found in the survey paper. Please click here to view the comprehensive archive of papers.
Timeline of representative models.
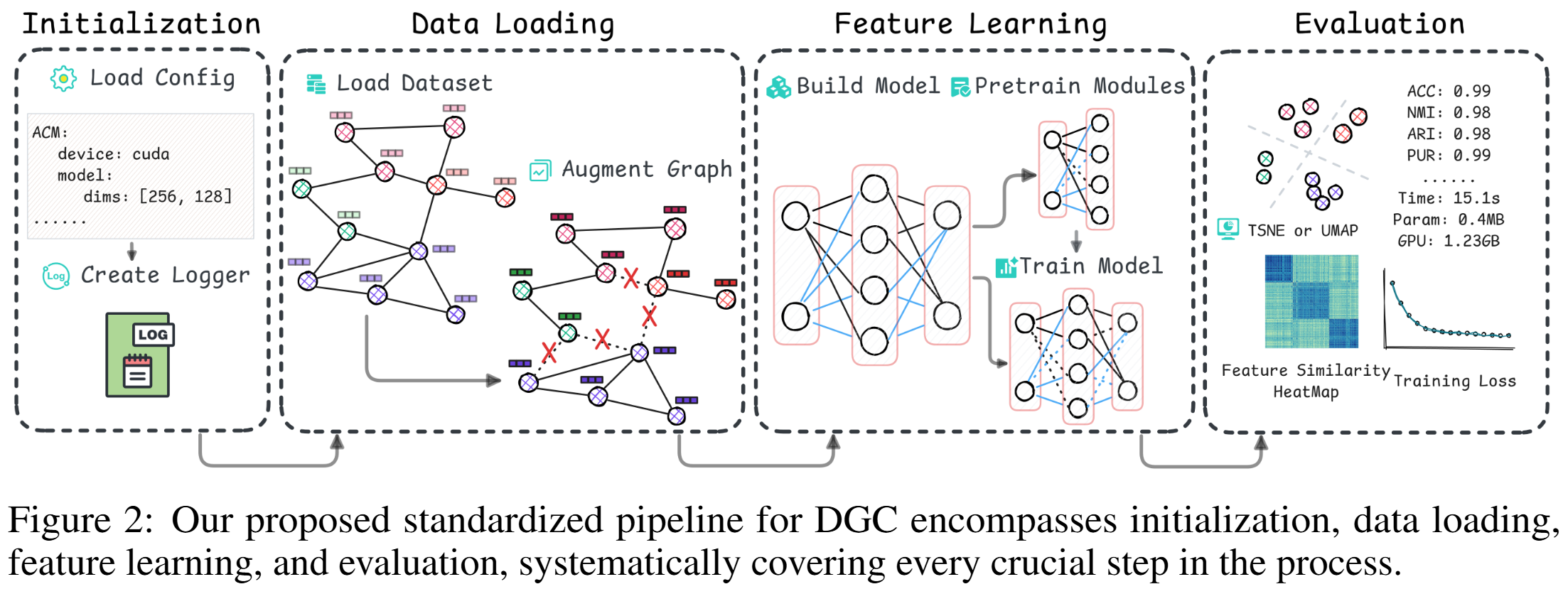
DGCBench encompasses 12 diverse datasets with different characteristics and 12 state-of-the-art methods from all major paradigms. By integrating them into a standardized pipeline, we ensure fair, reproducible, and comprehensive evaluations across multiple dimensions.
- Integration of multiple deep graph clustering models. Supported Models
- Support for various graph datasets from PyG and OGB. Supported Datasets
- Model evaluation and visualization capabilities.
- Standardized Pipeline.
It is recommended to use conda to create a virtual python environment.
conda create --name DGCbench python=3.8
conda activate DGCbenchIt is recommended to install GPU or CPU version of PyTorch according to the device in advance, version >=2.0.1.
-
(Plan 1) Install with Pip
pip install pydgc
-
(Plan 2) Installation for local development
git clone https://github.com/Marigoldwu/PyDGC.git cd PyDGC pip install -e .
Take GAE as an example. You can run the following command to reproduce the results of GAE on CORA dataset.
Create a folder dgc to store the results.
mkdir dgc
cd dgc
mkdir gae
cd gaeCopy config.yaml and run.py of GAE from our github repository to the current folder.
wget https://raw.githubusercontent.com/Marigoldwu/PyDGC/master/examples/pipelines/gae/config.yaml
wget https://raw.githubusercontent.com/Marigoldwu/PyDGC/master/examples/pipelines/gae/run.pyRun the pipeline.
python run.pyYou can also specify arguments in the command line:
python run.py --dataset_name CORA -eval_each --rounds 1Note:
--dataset_nameis the name of the dataset.--roundsis the number of times to run the pipeline.-eval_eachis the flag to evaluate the model after each epoch.
Other optional arguments:
--cfg_file_path YourPath # path of corresponding configurations file
--flag FlagContent # Descriptions
--drop_edge float # probability of dropping edges
--drop_feature float # probability of dropping features
--add_edge float # probability of adding edges
--add_noise float # standard deviation of Gaussian Noise
-pretrain # only run the pretraining stage in the modelfrom pydgc.models import DGCModel
class MyModel(DGCModel):
def __init__(self, logger, cfg):
super(MyModel).__init__(logger, cfg)
your_model = ... # Your model
self.loss_curve = []
self.nmi_curve = []
self.best_embedding = None
self.best_predicted_labels = None
self.best_results = {'ACC': -1}
def forward(self, data):
... # forward process
return something
# If needed
def loss(self, *args, **kwargs):
# If needed
def pretrain(self, data, cfg, flag):
def train_model(self, data, cfg, flag):
def get_embedding(self, data):
def clustering(self, data):
embedding = self.get_embedding(data)
# clustering
return embedding, labels_, clustering_centers
def evaluate(self, data):
embedding, predicted_labels, clustering_centers = self.clustering(data)
ground_truth = data.y.numpy()
metric = DGCMetric(ground_truth, predicted_labels.numpy(), embedding, data.edge_index)
results = metric.evaluate_one_epoch(self.logger, self.cfg.evaluate)
return embedding, predicted_labels, resultsfrom pydgc.pipelines import BasePipeline
from pydgc.utils import perturb_data
import MyModel # import your own model
class MyPipeline(BasePipeline):
def __init__(self, args):
super(MyPipeline).__init__(args)
def augmentation(self):
self.data = perturb_data(self.data, self.cfg.dataset.augmentation)
# other augmentations if needed
def build_model(self):
model = MyModel(self.logger, self.cfg)
self.logger.model_info(model)
return model| No. | Dataset | #Samples | #Features | #Edges | #Classes | Homo. Ratio |
|---|---|---|---|---|---|---|
| 1 | Wiki | 2,405 | 4,973 | 17,981 | 17 | 0.71 |
| 2 | Cora | 2,708 | 1,433 | 5,429 | 7 | 0.81 |
| 3 | ACM | 3,025 | 1,870 | 13,128 | 3 | 0.82 |
| 4 | Citeseer | 3,327 | 3,703 | 9,104 | 6 | 0.74 |
| 5 | DBLP | 4,057 | 334 | 3,528 | 4 | 0.80 |
| 6 | PubMed | 19,717 | 500 | 88,648 | 3 | 0.80 |
| 7 | Ogbn-arXiv | 169,343 | 128 | 2,315,598 | 40 | 0.65 |
| 8 | USPS(3NN) | 9,298 | 256 | 27,894 | 10 | 0.98 |
| 9 | HHAR(3NN) | 10,299 | 561 | 30,897 | 6 | 0.95 |
| 10 | BlogCatalog | 5,196 | 8,189 | 343,486 | 6 | 0.40 |
| 11 | Flickr | 7,575 | 12,047 | 479,476 | 9 | 0.24 |
| 12 | Roman-empire | 22,662 | 300 | 65,854 | 18 | 0.05 |
The 12 datasets above are benchmark datasets introduced in our paper. More Datasets will be introduced.
ADGC: Awesome-Deep-Graph-Clustering
Older version of this repository: A-Unified-Framework-for-Attribute-Graph-Clustering