Visual Assessment of Brain Registration Quality in Functional MRI studies
Several measures and tools have been proposed to asses quality control (QC) of functional magnetic resonance imaging (fMRI) data, most of them focus on the quality of raw data images out of the scanner (see Liu TT (2015) for a review). Very few of them focus on the QC of preprocessed fMRI data which are highly impacted by the typical image preprocessing steps (brain extraction and coregistration).
Studies that addresses QC of preprocessed data are divided in two categories; Manual QC and automated QC. This document describe the major steps to go through to manual QC of fMRI preprocessed data
For overall QC Status three values are acceptable:
-
Fail: A major flaw has been identified. Typically misregistration, artefacts or deformation.
-
Maybe: A minor problem has been identified, sometimes with the pipeline itself or in the raw data. Most common are brain extraction (BET) for structural images, Misregistration (MR) and misplaced field of view for functional images (FOV). Other less common causes of maybe are Brain Deformation (DEF), Brain Abnormality (ABN), Ghosthing (GHO), Motion artifacts (MOT), and, in some cases, Arterial artefacts (ART). Extreme variants of these issues may qualify for a Fail.
-
OK: Neither Fail nor Maybe. None or very minor MR, BET issues, and small FOV, DEF, ABN, GHO, MOT or ART. Issues such as Signal Loss (SL), Ventricule mis alligned (VENT) or brain atrophy (ATR) can be listed with an OK status, as they cannot be corrected fully by registration. However, once flagged, these issues can be used to build confound regressors in group analysis. An acceptable FOV will depend on the target coverage of the study (e.g. if cerebellum is excluded by design, this should not be flagged by FOV). Similarly, at 3T, normal DEF and SL in the temporal and orbitofrontal cortices will not be flagged, as they are observed systematically (unless a B0 field correction is implemented in the pipeline).
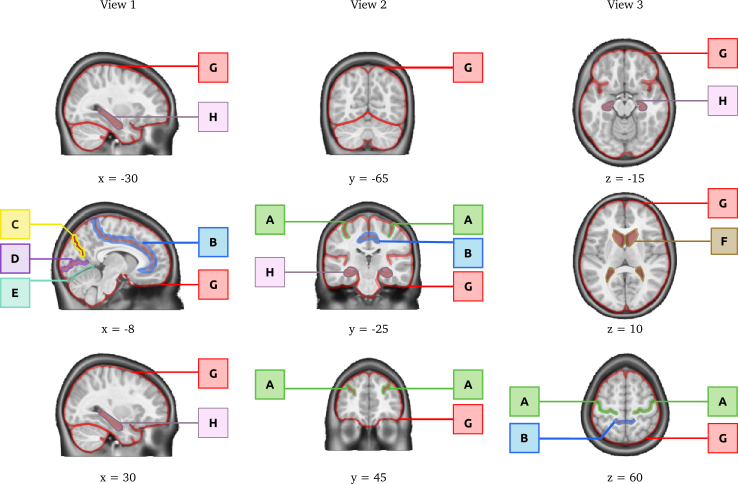
The anatomical landmarks that should be well aligned in a successful coregistration include: central sulcus (A), cingulate sulcus (B), parieto-occipital fissure (C), calcarine fissure (D), tentorium cerebellum (E), the lateral ventricles (F), the outline of the brain (G) and the hippocampal formation (H) bilateraly. The landmarks are outlined on an individual brain after successful non-linear coregistration in stereotaxic space.
The most frequent issues related to brain segmentation and spatial normalisation are listed below with figures for each Fail / Maybe / OK cases.